Spectral Models
Overview
Section titled “Overview”sensipy supports time-resolved spectral models that describe how the source emission evolves over time. The data should contain spectra at varying times (or lightcurves at varying energies, depending on how you prefer to think about it).
Currently, the supported file formats are:
- CSV files (recommended)
- FITS files
- Text files (directory of
.txtfiles)
Mock Data Files
Section titled “Mock Data Files”The package includes example mock data files that you can use for testing and learning. These files are installed with the package and can be accessed using the get_data_path() utility function:
Directorydata/
Directorymock_data/
- GRB_42_mock.csv (CSV format spectral model)
- GRB_42_mock.fits (FITS format spectral model)
- GRB_42_mock_metadata.csv (metadata for the source)
These example files demonstrate the expected format and can be used to test your setup. For real analysis, you’ll need to provide your own spectral model data.
CSV File Format
Section titled “CSV File Format”The code expects a CSV file with three columns: time, energy, and flux (dNdE) with the following units:
- time: seconds (s)
- energy: GeV
- dNdE: 1 / (cm² s GeV) - differential flux
Multiple spectra can be included in the same file, each with a different time. The code will automatically organize the data into a time-energy grid.
Sample CSV File
Section titled “Sample CSV File”A full sample CSV file is included with the package. The first rows look something like this:
time [s],energy [GeV],dNdE [cm-2 s-1 GeV-1]1.061,1.04823,1e-081.061,1.14937,8.3179e-091.061,1.26026,6.9185e-091.061,1.38184,5.7547e-091.061,1.51516,4.7861e-09...Column Name Flexibility
Section titled “Column Name Flexibility”The CSV reader is flexible with column names:
- Column names are case-insensitive
- Substring matching is supported (e.g., “Time” will match “time [s]”)
- The code looks for columns containing:
time,energy, andfluxordnde
Loading a CSV File
Section titled “Loading a CSV File”from sensipy.source import Sourcefrom sensipy.util import get_data_pathimport astropy.units as u
# Get path to package mock datamock_data_path = get_data_path("mock_data/GRB_42_mock.csv")
# Load CSV spectral modelsource = Source( filepath=str(mock_data_path), min_energy=30 * u.GeV, max_energy=10 * u.TeV,)
# Optionally add EBL absorptionsource_with_ebl = Source( filepath=str(mock_data_path), min_energy=30 * u.GeV, max_energy=10 * u.TeV, ebl="franceschini" z=0.5,)FITS File Format
Section titled “FITS File Format”FITS files can also be used to store spectral models. The expected structure is:
- HDU[1]: Energy array (in GeV)
- HDU[2]: Time array (in seconds)
- HDU[3]: Flux array (2D, shape:
[n_energy, n_time], units: cm⁻² s⁻¹ GeV⁻¹) - HDU[0] (optional): Metadata (source properties)
FITS Example
Section titled “FITS Example”from sensipy.source import Sourcefrom sensipy.util import get_data_pathimport astropy.units as u
# Get path to package mock datamock_fits_path = get_data_path("mock_data/GRB_42_mock.fits")
# Load FITS spectral modelsource = Source( filepath=str(mock_fits_path), min_energy=30 * u.GeV, max_energy=10 * u.TeV, ebl="dominguez", z=0.5,)Metadata
Section titled “Metadata”Metadata provides additional information about the source, such as sky coordinates, distance, energy output, and jet properties.
User-Defined Metadata
Section titled “User-Defined Metadata”sensipy uses a completely user-defined metadata system. There are no built-in metadata fields—you define whatever metadata keys you need for your analysis. Metadata is stored in a dictionary and can be accessed via attribute notation (similar to pandas DataFrame columns).
CSV Metadata File
Section titled “CSV Metadata File”For CSV spectral models, you can provide a separate metadata file with the same basename plus _metadata.csv:
GRB_42_mock.csvGRB_42_mock_metadata.csvThe metadata CSV file should have columns: parameter, value, and optionally units:
parameter,value,unitsevent_id,42.0,longitude,0.0,radlatitude,1.0,raddistance,100000.0,kpcAny parameter names you include will be stored in the metadata dictionary. The units column is optional—if provided, values will be converted to astropy Quantity objects with the specified units.
FITS Metadata
Section titled “FITS Metadata”For FITS files, metadata can be included in the header of HDU[0] using a flexible format. sensipy reads any non-standard FITS header keys and converts them to metadata.
FITS Header Format
Section titled “FITS Header Format”The FITS header format uses a comment field to specify the metadata slug and optional unit:
from astropy.io import fits
header = fits.Header()# Format: header["FITS_KEY"] = (value, "slug [unit]")header["EVENT_ID"] = (42, "event_id")header["LONG"] = (0.0, "longitude [rad]")header["LAT"] = (1.0, "latitude [rad]")header["DISTANCE"] = (100000.0, "distance [kpc]")header["EISO"] = (2e50, "eiso [erg]")
# Empty values are ignoredheader["AUTHOR"] = ("", "author") # This will be skipped
# Keys without comments use the header key name (lowercase) as the slugheader["CUSTOM_FIELD"] = (123.45, "") # Stored as "custom_field"Format rules:
- Value: The actual metadata value (number or string)
- Comment: Format
"slug [unit]"where:slugis the metadata key name (will be converted to lowercase, spaces/special chars become underscores)[unit]is optional - if provided, the value is converted to anastropy.units.Quantityorastropy.coordinates.Distanceobject
- Empty values: Header entries with empty string values are ignored
- No comment: If no comment is provided, the header key name (lowercase) is used as the slug
Special handling:
- Keys named
distanceordistwith units are converted toastropy.coordinates.Distanceobjects - Standard FITS keywords (like
SIMPLE,BITPIX,NAXIS, etc.) are automatically skipped
Example:
from sensipy.source import Sourcefrom sensipy.util import get_data_path
# Load FITS file with flexible metadatamock_fits_path = get_data_path("mock_data/GRB_42_mock.fits")source = Source(mock_fits_path)
# Access metadata via attribute notationprint(f"Event ID: {source.event_id}") # 42print(f"Longitude: {source.longitude}") # 0.0 radprint(f"Latitude: {source.latitude}") # 1.0 radprint(f"Distance: {source.distance}") # 100000.0 kpc (Distance object)Accessing Metadata
Section titled “Accessing Metadata”Once loaded, metadata can be accessed in two ways:
- Via attribute notation (recommended, similar to pandas):
from sensipy.source import Sourcefrom sensipy.util import get_data_path
mock_data_path = get_data_path("mock_data/GRB_42_mock.csv")source = Source(filepath=str(mock_data_path))
# Access metadata via attributes (using keys from your metadata file)print(f"Event ID: {source.event_id}")print(f"Distance: {source.distance}")print(f"Coordinates: RA={source.longitude}, Dec={source.latitude}")
# Note: Only keys present in your metadata file are available# The example above uses keys from the mock data (event_id, longitude, latitude, distance)- Via the metadata dictionary:
# Access the full metadata dictionarymeta = source.metadataprint(f"All metadata keys: {list(meta.keys())}")print(f"Event ID: {meta.get('event_id')}")print(f"Distance: {meta.get('distance')}")Setting Custom Metadata
Section titled “Setting Custom Metadata”You can add or modify metadata at any time:
# Set custom metadata fieldssource.my_custom_field = "some value"source.another_field = 123.45
# Access them via attribute notationprint(source.my_custom_field) # "some value"print(source.another_field) # 123.45
# Or via the metadata dictionaryprint(source.metadata['my_custom_field'])Text File Directory Format
Section titled “Text File Directory Format”For legacy support, sensipy can also read spectral data from a directory of text files. Each file should be named with the pattern:
{basename}_tobs=NN.txtWhere NN is the observation time index. Each file contains two columns: energy (GeV) and flux (cm⁻² s⁻¹ GeV⁻¹).
Spectral Grid and Interpolation
Section titled “Spectral Grid and Interpolation”After loading a spectral model, sensipy creates an interpolation grid in log-space (log energy, log time) to enable efficient querying at arbitrary times and energies.
Querying the Spectrum
Section titled “Querying the Spectrum”import astropy.units as u
# Get spectrum at a specific timetime = 100 * u.sspectrum = source.get_spectrum(time=time)
# Get flux at a specific energy and timeenergy = 1 * u.TeVflux = source.get_flux(energy=energy, time=time)
# Get lightcurve at a specific energylightcurve = source.get_flux(energy=energy)The following plots demonstrate these querying methods using the mock data:
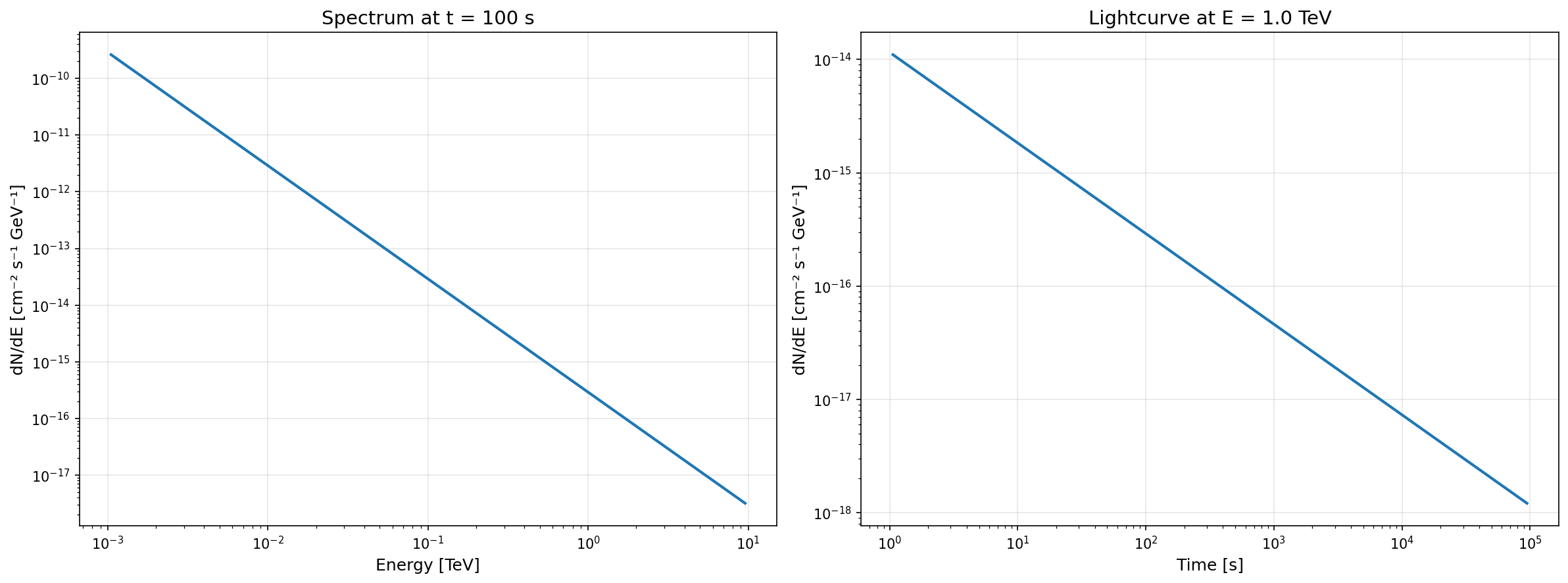
Visualizing the Spectral Pattern
Section titled “Visualizing the Spectral Pattern”The spectral pattern is a 2D plot of the spectral energy distribution (SED) as a function of time and energy.
# Show the full time-energy spectral patternsource.show_spectral_pattern( resolution=100, # Grid resolution return_plot=True)Spectral pattern visualization:
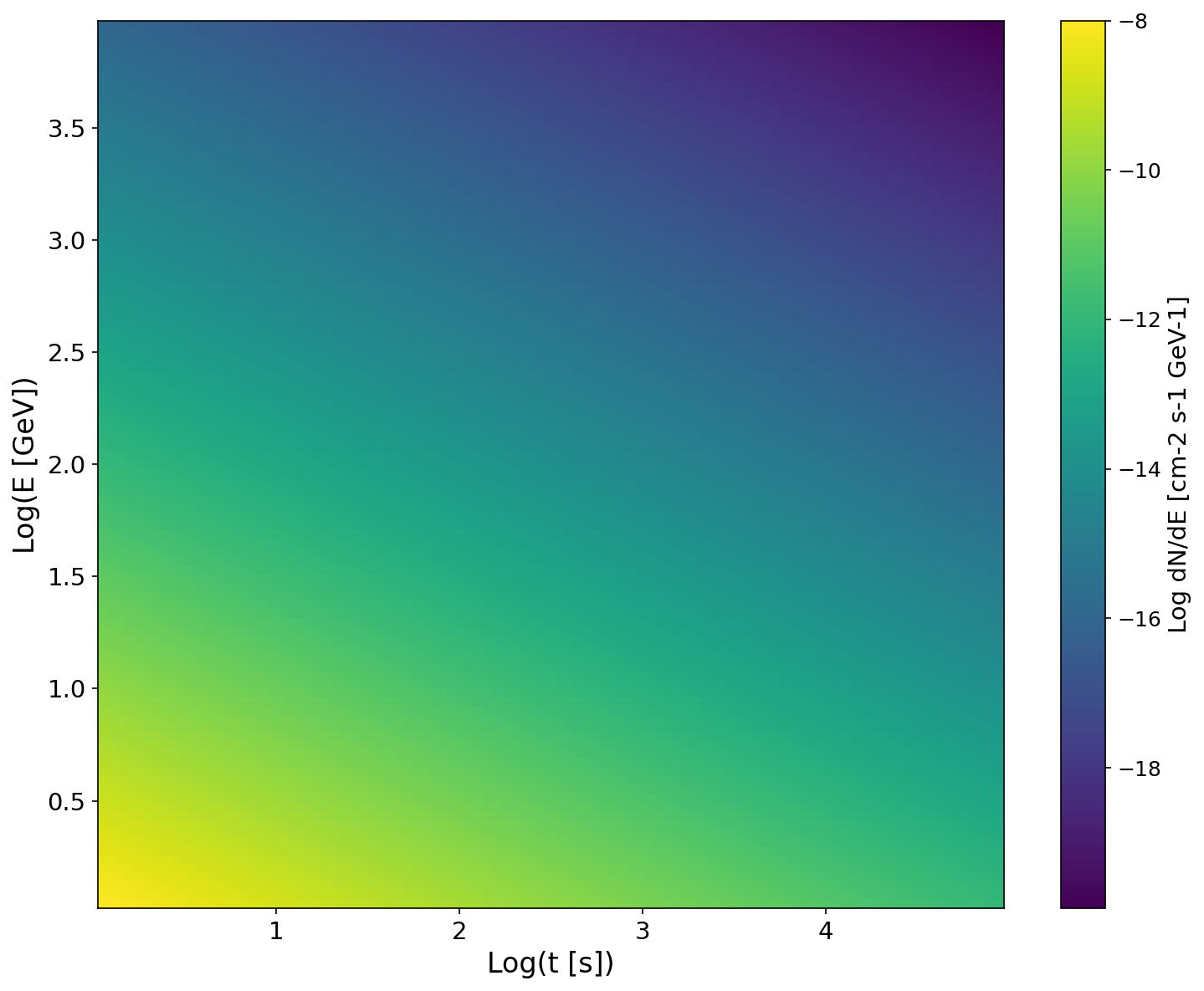
Setting Your Distance
Section titled “Setting Your Distance”For extragalactic sources, you need to specify the distance (or redshift) to apply EBL absorption correctly. You can set the distance either during initialization or after creating the Source object.
During Initialization
Section titled “During Initialization”You can provide either a redshift (z) or a distance (Distance object) when creating the Source:
from sensipy.source import Sourcefrom sensipy.util import get_data_pathimport astropy.units as u
mock_data_path = get_data_path("mock_data/GRB_42_mock.csv")
# Set redshift during initializationsource = Source( filepath=str(mock_data_path), min_energy=20 * u.GeV, max_energy=10 * u.TeV, z=0.5, # Redshift ebl="franceschini",)from sensipy.source import Sourcefrom sensipy.util import get_data_pathfrom astropy.coordinates import Distanceimport astropy.units as u
mock_data_path = get_data_path("mock_data/GRB_42_mock.csv")
# Set distance during initializationdistance = Distance(3000 * u.Mpc)source = Source( filepath=str(mock_data_path), min_energy=20 * u.GeV, max_energy=10 * u.TeV, distance=distance, # Distance object ebl="franceschini",)Note: Only one of distance or z can be provided, not both. If both are provided, a ValueError will be raised.
After Initialization
Section titled “After Initialization”You can also set or update the distance using the set_ebl_model() method:
from sensipy.source import Sourcefrom sensipy.util import get_data_pathimport astropy.units as u
mock_data_path = get_data_path("mock_data/GRB_42_mock.csv")
# Create source without distancesource = Source( filepath=str(mock_data_path), min_energy=20 * u.GeV, max_energy=10 * u.TeV,)
# Set distance and EBL model latersource.set_ebl_model("franceschini", z=0.5)from sensipy.source import Sourcefrom sensipy.util import get_data_pathfrom astropy.coordinates import Distanceimport astropy.units as u
mock_data_path = get_data_path("mock_data/GRB_42_mock.csv")
# Create source without distancesource = Source( filepath=str(mock_data_path), min_energy=20 * u.GeV, max_energy=10 * u.TeV,)
# Set distance and EBL model laterdistance = Distance(3000 * u.Mpc)source.set_ebl_model("franceschini", distance=distance)Distance Override Priority
Section titled “Distance Override Priority”If you provide distance or z during initialization, it will override any distance information extracted from the metadata file. The priority order is:
- Explicit
distanceorzparameter inSource()(highest priority) - Metadata distance from CSV or FITS file
- No distance - EBL won’t be applied unless distance is set later
Using Your Own Data
Section titled “Using Your Own Data”To use your own spectral models:
- Prepare your data in CSV or FITS format (see format details above)
- Include metadata (optional but recommended) for source properties like distance, coordinates, etc.
- Load the data using the
Sourceclass
from sensipy.source import Sourceimport astropy.units as u
# Load your custom spectral modelsource = Source( filepath="path/to/your/model.csv", min_energy=30 * u.GeV, max_energy=10 * u.TeV, ebl="franceschini", # Optional: add EBL absorption for extragalactic sources z=0.5,)Exporting to Gammapy Spectral Models
Section titled “Exporting to Gammapy Spectral Models”sensipy can export spectra as Gammapy spectral models for use in likelihood fitting, sensitivity calculations, and other Gammapy workflows. Two types of models are available:
Power Law Approximation
Section titled “Power Law Approximation”The get_powerlaw_spectrum() method fits a power law to the spectrum at a given time and returns a PowerLawSpectralModel:
from sensipy.source import Sourcefrom sensipy.util import get_data_pathimport astropy.units as u
mock_data_path = get_data_path("mock_data/GRB_42_mock.csv")source = Source( filepath=str(mock_data_path), min_energy=20 * u.GeV, max_energy=10 * u.TeV,)
time = 100 * u.spowerlaw_model = source.get_powerlaw_spectrum(time)
# Use in Gammapy workflowsfrom gammapy.modeling.models import SkyModelsky_model = SkyModel(spectral_model=powerlaw_model)Template Spectrum
Section titled “Template Spectrum”The get_template_spectrum() method extracts the full energy spectrum at a given time and returns a ScaledTemplateModel:
time = 100 * u.stemplate_model = source.get_template_spectrum(time, scaling_factor=1.0)
# Template models preserve the full spectral shape# Useful when power law approximation is insufficientEBL Absorption
Section titled “EBL Absorption”Both methods support automatic EBL absorption via the use_ebl parameter:
source = Source( filepath=str(mock_data_path), min_energy=20 * u.GeV, max_energy=10 * u.TeV, z=1.0, ebl="franceschini", # Set EBL model)
time = 100 * u.s# Apply EBL absorption automatically (or use default: use_ebl=None)powerlaw_with_ebl = source.get_powerlaw_spectrum(time, use_ebl=True)source = Source( filepath=str(mock_data_path), min_energy=20 * u.GeV, max_energy=10 * u.TeV, z=1.0, ebl="franceschini", # Set EBL model)
time = 100 * u.s# Apply EBL absorption automatically (or use default: use_ebl=None)template_with_ebl = source.get_template_spectrum(time, use_ebl=True)Comparison plots:
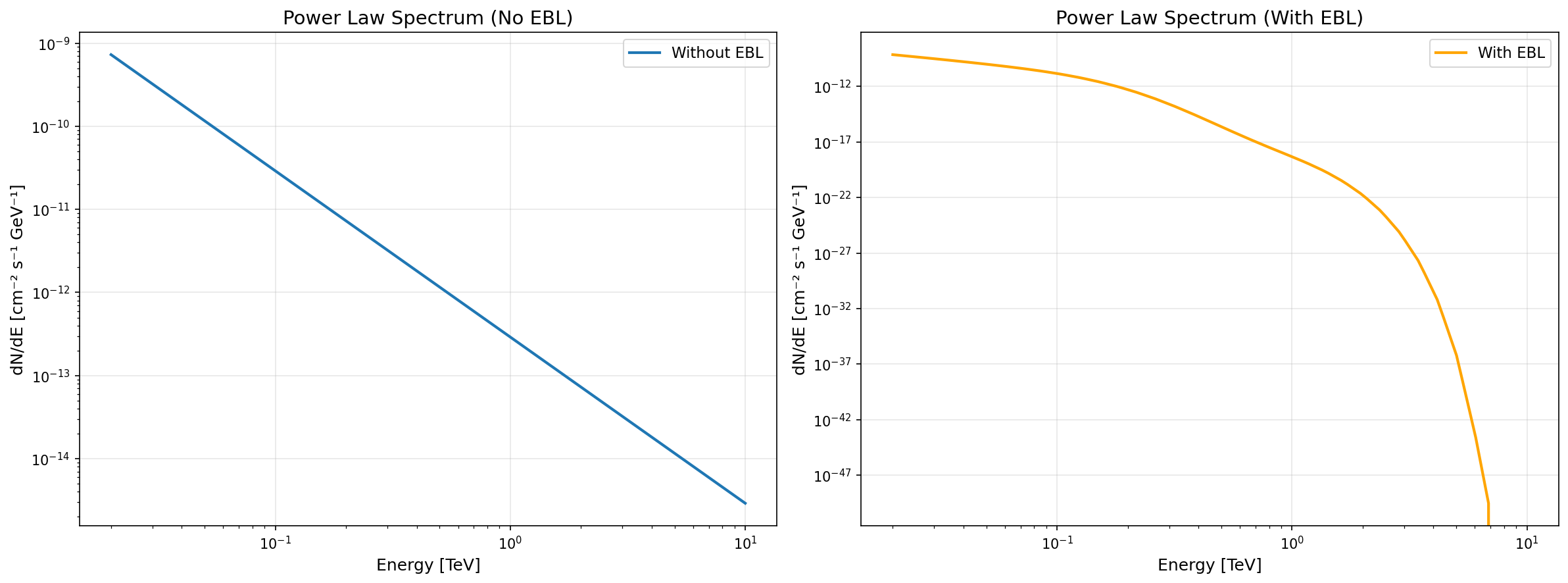
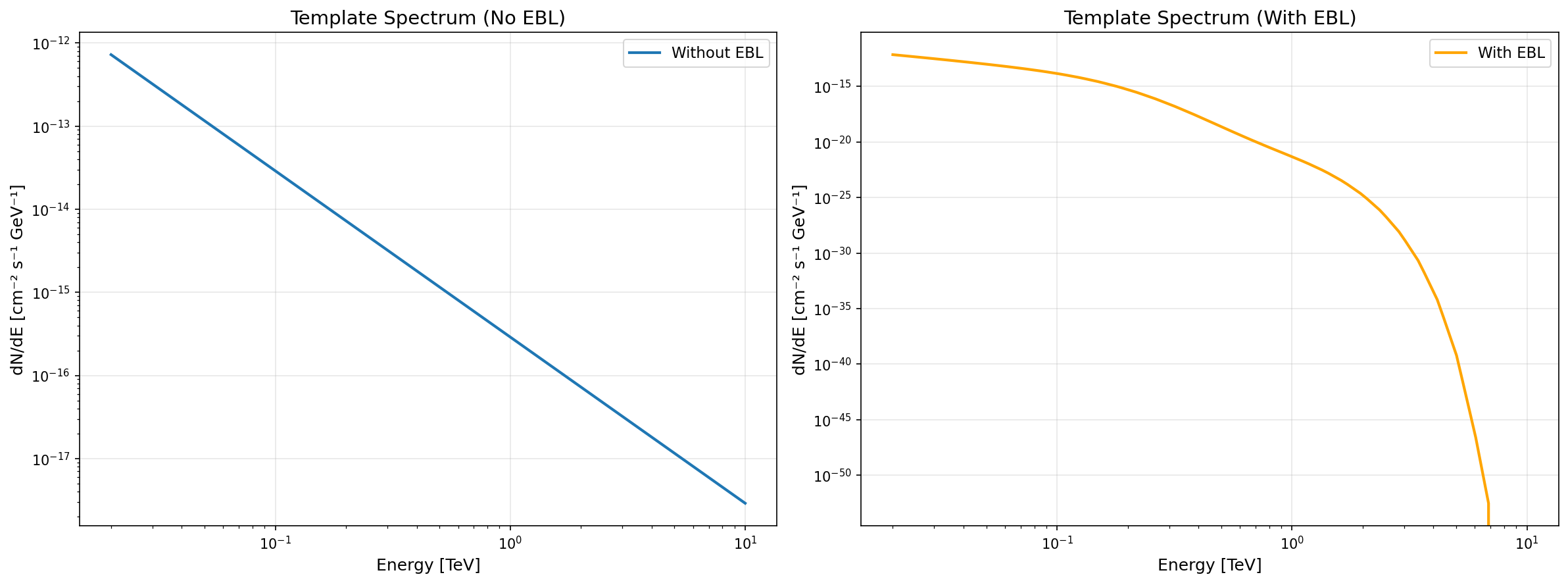
Automatic Energy Range in Plots
Section titled “Automatic Energy Range in Plots”Both exported models have an overridden .plot() method that automatically uses the source’s min_energy and max_energy:
# Plot automatically uses source.min_energy and source.max_energypowerlaw_model.plot()
# You can still override the energy range if neededpowerlaw_model.plot(energy_range=(0.1 * u.TeV, 5 * u.TeV))Energy Range Considerations
Section titled “Energy Range Considerations”When loading spectral models, you should specify the energy range of interest. This helps:
- Optimize performance by limiting interpolation to relevant energies
- Match observatory capabilities (e.g., CTA energy range)
- Ensure EBL absorption is applied correctly
# Typical CTA energy rangemin_energy = 20 * u.GeV # or 0.02 TeVmax_energy = 10 * u.TeV
source = Source( filepath="path/to/model.csv", min_energy=min_energy, max_energy=max_energy,)